About inMOTIFin
inMOTIFin is a lightweight python package for simulating and modifying cis-regulatory elements. For the preprint detailing its implementation and use cases, visit https://doi.org/10.48550/arXiv.2506.20769. It can be used directly on the JASPAR website from the 2026 release, as well as a command line tool and as a Python module, with the latter providing more control over the simulation. The basic blocks of the simulation are the motifs and the backgrounds. A final sequence will include one or more motifs inserted into the background sequence. More than 40 user defined parameters can control the possibilities of these insertions and through sampling inMOTIFin realises actual motif-in-sequences.
Usage on the JASPAR website
inMOTIFin is available as a cart-related tool on the JASPAR website.
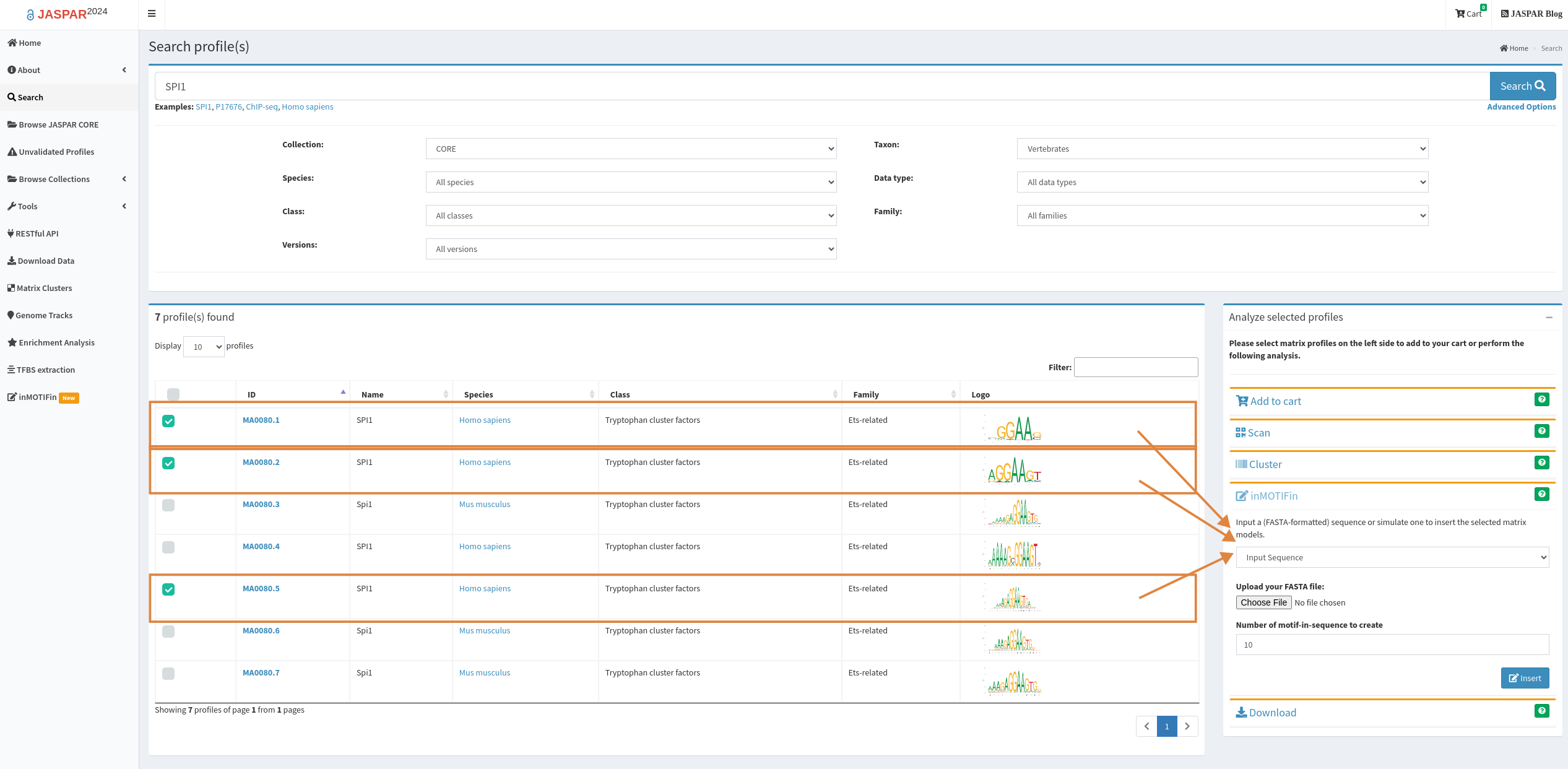
Motifs are loaded directly upon selection from the available profiles. Backgrounds can be uploaded as fasta files or simulated by setting the parameters for the number and length of backgrounds, the alphabet, and the probability of each letter in the alphabet. Note: this should sum to 1.

Upon running the tool via the "insert" button, the results appear in a new window.
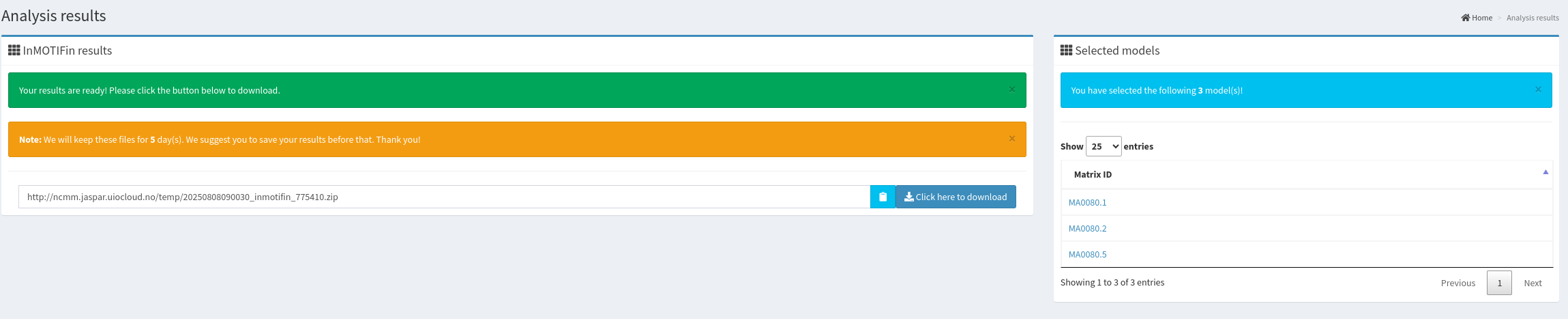
The content of the downloaded folder (after unzipping) is a set of files.
- inmotifin_final_sequences.fa is a fasta file containing all the motif-in-sequences. The name of the sequences are the index_title_backgroundID.
- inmotifin_probabilistic_final_sequences.fa is a numpy dictionary file containing all the per position letter probabilities of the motif-in-sequences.
- inmotifin_inserted_instances.bed is a bed file containing the locations of the motifs in each of the motif-in-sequences. The first column is the name of the sequence as in the fasta file: index_title_backgroundID. The second and third columns are the start and end coordinates of the inserted motif instance. The fourth column is the name of the inserted instance: title_motifID_instance. The score column is ".". The strand column is + or - depending on the orientation of the motif instance.
- inmotifin_occurrence_summaries.json is json file with counts for the values of all nodes. Number of times each background, group, motif, and orientation was selected. The number of occurrences of specific motif instances (note that the background might include more such instances not controlled by actual insertion). The number of how many times a specific number of instances per sequences was selected (here set to 1). Number of selected start positions (here set to central), the occurrences of different motif lengths, and the number of backgrounds without motif inserted in them (here set to 0).
- inmotifin_backgrounds.fa is a fasta file containing all the background sequences.
- inmotifin_background_probabilities.npz is a numpy dictionary file containing all the per position letter probabilities of the background sequences.
- inmotifin_dagsim_table.csv is a table produced by DagSim framework, including all values of all nodes for each simulation round.
- inmotifin_group_frequency.tsv is the occurence frequency or selection probability of each group. Here only one group is set.
- inmotifin_group_group_transition_probabilities.tsv is the transition probability of each group pairs in the Markov chain. Here not applicable because only one group is set.
- inmotifin_motif_group_membership.tsv is a tsv file where the first column is the group IDs, and the second column lists the motfIDs that are assigned to the corresponding group. Here only one group is set.
- inmotifin_motif_freq_per_group.tsv is a table exported from pandas showing the probability of selection of each motif from each group. Here equal probability is set.
Local installation of the tool
inMOTIFin is available at the Python package registry: https://pypi.org/project/inMOTIFin/ as well as on DockerHub: https://hub.docker.com/r/cbgr/inmotifin.
Local usage of the tool
The main functionality of the tool can be run with the following command:
python -m inmotifin motif-in-seq --title simulation --num_motif_in_seq 100 --seed 47
where the highlighted options are:
- title is a string which will be the name of the output folder and the prefix of all generated output files.
- num_motif_in_seq defines how many sequences with inserted motif instances will be produced by inMOTIFin.
- seed is for reproducibility. In this specific case when no other parameters are set, both the backgrounds and the motifs are simulated. The sampling steps are always probabilistic, so it is a good idea to set a seed.
Motifs and backgrounds can be loaded from files, or created by simulation following user defined parameters. inMOTIFin supports the following file formats for motifs: meme, jaspar, or csv with a list of JASPAR matrix ids; and fasta files for backgrounds. Alternatively, motifs can be simulated from parameters such as alphabet, length, and letter-specific prior information about information content and occurrence probability parameterized by alpha values for a Dirichlet distribution. Background simulation can be position-independent sampling from a distribution when the alphabet and the probability of each letter in the alphabet are provided. Alternatively, a hidden Markov model can be fitted to user provided sequences (in fasta format) from which new backgrounds can be sampled.
A full list of optional parameters can be read at https://inmotifin.readthedocs.io/.
Other supported functionalities are:
$ python -m inmotifin Usage: python -m inmotifin [OPTIONS] COMMAND [ARGS]... You are running inMOTIFin Options: --help Show this message and exit. Commands: motif-in-seq Simulating sequences with inserted motif instances motifs Creating motifs given information content, length,... multimers Multimerizing motifs given motifs and distances random-sequences Creating random sequences given length and alphabet,...
With the "multimers" option, any set of motifs can be “multimerized” prior to insertion. This means that by providing a list of motif ids and a list of distances between consecutive ones (which may be negative), inMOTIFin can join motifs into multimers (i.e. consecutive sets of motifs with fixed grammar).